Navigation auf uzh.ch
Navigation auf uzh.ch
The mammalian circadian clock is composed molecularly of interlocking feedback loops of gene expression. While the epistatic relationship of the components within these loops is clear, the mechanism of action of many actors remains largely unknown. For example, within the principal feedback loop, although Period proteins are known to interact with Cryptochrome proteins and with CLOCK and BMAL1, the mechanism by which they repress clock gene transcription remains poorly understood (reviewed in (Albrecht and Eichele, 2003). Equally enigmatic is the question of how their repressive action is delayed relative to their expression at the RNA level in order to make a 24-hour clock, a question partly resolved by extensive post-translational modifications and their regulation (reviewed in Harms et al., 2004). To better understand the function of these proteins, we have taken a biochemical approach to look for interacting factors that might give clues to Period protein function. Recently, we have purified a PER1-containing complex and identified its other components: clock proteins CRY1 and CRY2, as well as two novel components, NONO and WDR5 (Brown et al, 2005b).
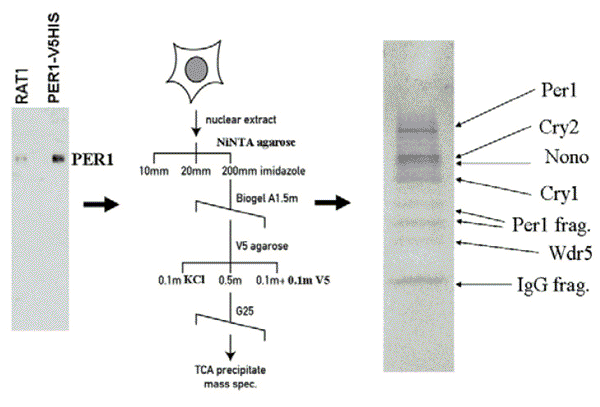
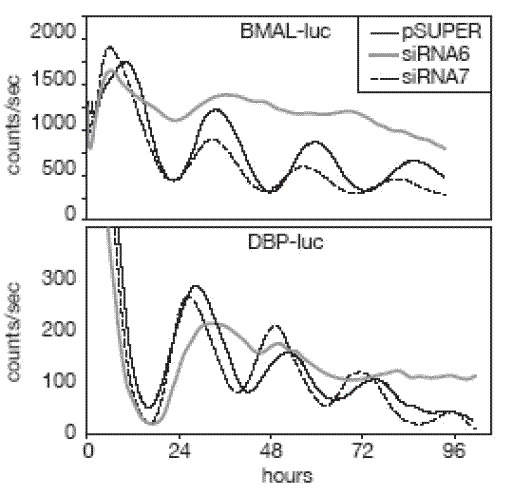
While the former is an adapter protein for a functionally well-characterized family of histone methylases involved in the regulation of tanscription, the latter has been found in a variety of contexts involving RNA transport and processing (Shav-Tal and Zipori, 2002). We have demonstrated by both gain- and loss-of-function experiments that WDR5 augments PERIOD protein-mediated transcriptional repression and alters circadian histone methylation at a clock-controlled gene. Using RNAi strategies in mammalian cells, we also showed that reduced levels of NONO result in shortened clock period length and attenuated circadian rhythms.
In collaboration with the Rosbash laboratory at Brandeis University, we learned that Drosophila homozygous for a hypomorphic allele of the homologous gene, NonA, are hyperactive and nearly arrhythmic. Surprisingly, in mammals NONO seems to interact with Period proteins to antagonize the negative limb of the circadian oscillator. Though NONO was associated with PER1, a repressor of circadian gene expression, its depletion resulted in the further repression of Period target loci. Thus, the PERIOD protein population is associated with both positive and negative factors that contribute to the regulation of circadian cycles (Brown et al., 2005b). We speculate that such dynamic regulation of Period protein function might add resilience to the circadian clock during perturbations in temperature and during cell division.
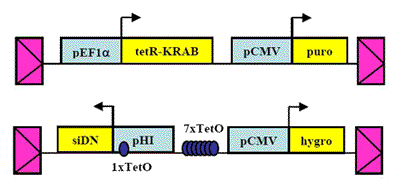
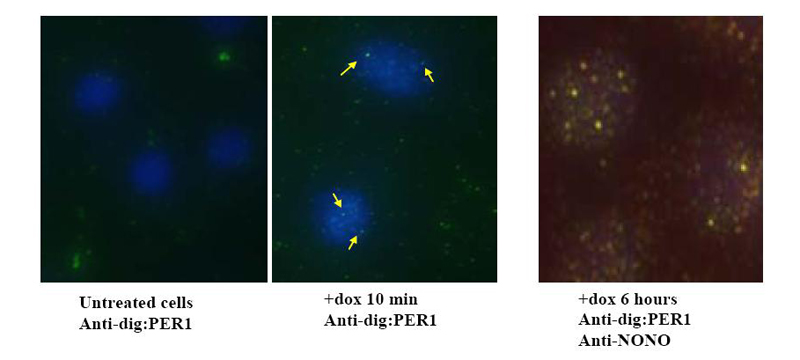
The system that we have begun to dissect possesses great richness as a paradigm for gene expression. Cyclic methylation of chromatin mediated by WDR5 occurs in the absence of cell division, suggesting a role for histone exchange factors or long sought demethylases. Indeed, further research has shown that PER-regulated genes undergo daily changes from euchromatin to facultative heterochromatin and back, thereby potentiating circadian gene expression (Ripperger and Schibler, 2006). Equally interesting, NONO has been characterized by several other labs, who have assigned it very diverse functions. Most of these functions are related to the transport and processing of RNA. Consistent with this hypothesis, the protein has two RNA-binding domains. It has been found as a member of a splicing complex and an RNA transport complex, and is an abundant component of paraspeckles, nuclear RNA retention bodies. These bodies were recently shown to be involved in the regulation of gene expression by conditional nuclear retention of transcripts (Prasanth et al., 2005). In the regulation of transcription, NONO has been found to be present at the promoter of repressed steroid receptor-regulated genes in ligand-dependent fashion (Mathur et al., 2001). We are currently taking various approaches to understanding the way in which NONO functions both within the circadian oscillator and more generally. These include loss-of-function experiments involving cell lines that reversibly repress the expression of the NONO gene in doxycycline-inducible fashion via siRNA expression, as well as genetically engineered mice harboring disruptions of the NONO locus. We are also actively characterizing the cell biology of the circadian clock and the role of NONO within it via immunohistochemical approaches and confocal microscopy.
________
Albrecht, U., and Eichele, G. (2003). The mammalian circadian clock. Curr Opin Genet Dev 13:271-277.
Brown, S. A., Ripperger, J., Kadener, S., Fleury-Olela, F., Vilbois, F., Rosbash, M., and Schibler, U. (2005b). PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308: 693-696.
Gribnau, J., de Boer, E., Trimborn, T., Wijgerde, M., Milot, E., Grosveld, F., and Fraser, P. (1998). Chromatin interaction mechanism of transcriptional control in vivo. Embo J 17: 6020-6027.
Harms, E., Kivimae, S., Young, M. W., and Saez, L. (2004). Posttranscriptional and posttranslational regulation of clock genes. J Biol Rhythms 19:361-373.
Mathur, M., Tucker, P. W., and Samuels, H. H. (2001). PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol 21:2298-2311.
Prasanth, K. V., Prasanth, S. G., Xuan, Z., Hearn, S., Freier, S. M., Bennett, C. F., Zhang, M. Q., and Spector, D. L. (2005). Regulating gene expression through RNA nuclear retention. Cell 123:249-263.
Ripperger, J. A., and Schibler, U. (2006). Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet., in press.
Shav-Tal, Y., and Zipori, D. (2002). PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett 531: 109-114.